本帖最后由 levycui 于 2018-7-31 10:54 编辑
问题导读:
1、如何使用支持向量机拟合线性回归?
2、TF中如何实现损失函数?
3、如何在TensorFlow中使用内核?
4、如何在系统中创建一个预测点网格?
上一篇:TensorFlow ML cookbook 第四章1节 应用线性SVM
减少到线性回归
支持向量机可用于拟合线性回归。 在本章中,我们将探讨如何使用TensorFlow执行此操作。
做好准备
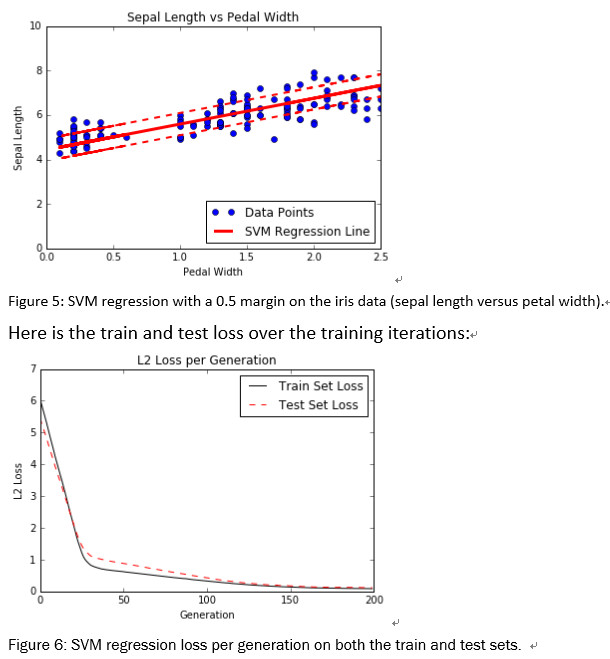
可以将相同的最大边际概念应用于拟合线性回归。 我们可以考虑最大化包含最多(x,y)点的边距,而不是最大化分隔类的边距。 为了说明这一点,我们将使用相同的虹膜数据集,并表明我们可以使用此概念来拟合萼片长度和花瓣宽度之间的线。
相应的损失函数将类似于max
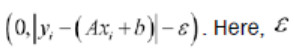 这里是边距宽度的一半,如果一个点位于该区域,则损失等于零。
这里是边距宽度的一半,如果一个点位于该区域,则损失等于零。
怎么做
1.首先,我们加载必要的库,启动图表,然后加载虹膜数据集。 之后,我们将数据集拆分为训练集和测试集,以显示两者的损失。 使用以下代码:
[mw_shl_code=python,true]import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn import datasets
sess = tf.Session()
iris = datasets.load_iris()
x_vals = np.array([x[3] for x in iris.data])
y_vals = np.array([y[0] for y in iris.data])
train_indices = np.random.choice(len(x_vals), round(len(x_ vals)*0.8), replace=False)
test_indices = np.array(list(set(range(len(x_vals))) - set(train_ indices)))
x_vals_train = x_vals[train_indices]
x_vals_test = x_vals[test_indices]
y_vals_train = y_vals[train_indices]
y_vals_test = y_vals[test_indices][/mw_shl_code]
对于此示例,我们将数据拆分为训练和测试。 将数据拆分为三个数据集也很常见,其中包括验证集。 我们可以使用此验证集来验证我们在训练它们时不会过度拟合模型。
2.让我们声明我们的批量大小,占位符和变量,并创建我们的线性模型,如下所示:
[mw_shl_code=python,true]batch_size = 50
x_data = tf.placeholder(shape=[None, 1], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
A = tf.Variable(tf.random_normal(shape=[1,1]))
b = tf.Variable(tf.random_normal(shape=[1,1]))
model_output = tf.add(tf.matmul(x_data, A), b) [/mw_shl_code]
3.现在我们宣布我们的损失函数。 如前文所述,实现了损失函数。 请记住,epsilon是我们的损失函数的一部分,它允许软边距而不是硬边距。
[mw_shl_code=python,true]epsilon = tf.constant([0.5])
loss = tf.reduce_mean(tf.maximum(0., tf.sub(tf.abs(tf.sub(model_ output, y_target)), epsilon))) [/mw_shl_code]
4.我们创建一个优化器并接下来初始化我们的变量,如下所示:
[mw_shl_code=python,true]my_opt = tf.train.GradientDescentOptimizer(0.075)
train_step = my_opt.minimize(loss)
init = tf.initialize_all_variables()
sess.run(init) [/mw_shl_code]
5.现在我们迭代200次训练迭代并保存训练和测试损失以便以后绘图:
[mw_shl_code=python,true]train_loss = []
test_loss = []
for i in range(200):
rand_index = np.random.choice(len(x_vals_train), size=batch_ size)
rand_x = np.transpose([x_vals_train[rand_index]])
rand_y = np.transpose([y_vals_train[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
temp_train_loss = sess.run(loss, feed_dict={x_data: np.transpose([x_vals_train]), y_target: np.transpose([y_vals_ train])})
train_loss.append(temp_train_loss)
temp_test_loss = sess.run(loss, feed_dict={x_data: np.transpose([x_vals_test]), y_target: np.transpose([y_vals_ test])})
test_loss.append(temp_test_loss)
if (i+1)%50==0:
print('-----------')
print('Generation: ' + str(i))
print('A = ' + str(sess.run(A)) + ' b = ' + str(sess. run(b)))
print('Train Loss = ' + str(temp_train_loss))
print('Test Loss = ' + str(temp_test_loss)) [/mw_shl_code]
6.这导致以下输出:
[mw_shl_code=python,true]Generation: 50
A = [[ 2.20651722]] b = [[ 2.71290684]]
Train Loss = 0.609453
Test Loss = 0.460152
-----------
Generation: 100
A = [[ 1.6440177]] b = [[ 3.75240564]]
Train Loss = 0.242519
Test Loss = 0.208901
-----------
Generation: 150
A = [[ 1.27711761]] b = [[ 4.3149066]]
Train Loss = 0.108192
Test Loss = 0.119284
-----------
Generation: 200
A = [[ 1.05271816]] b = [[ 4.53690529]]
Train Loss = 0.0799957
Test Loss = 0.107551 [/mw_shl_code]
7.我们现在可以提取我们找到的系数,并获得最佳拟合线的值。 出于绘图目的,我们也将获得边距的值。 使用以下代码:
[mw_shl_code=python,true][[slope]] = sess.run(A)
[[y_intercept]] = sess.run(b)
[width] = sess.run(epsilon)
best_fit = []
best_fit_upper = []
best_fit_lower = []
for i in x_vals:
best_fit.append(slope*i+y_intercept)
best_fit_upper.append(slope*i+y_intercept+width)
best_fit_lower.append(slope*i+y_intercept-width) [/mw_shl_code]
8.最后,这里是用拟合线和列车测试损失绘制数据的代码:
[mw_shl_code=python,true]plt.plot(x_vals, y_vals, 'o', label='Data Points')
plt.plot(x_vals, best_fit, 'r-', label='SVM Regression Line', linewidth=3)
plt.plot(x_vals, best_fit_upper, 'r--', linewidth=2)
plt.plot(x_vals, best_fit_lower, 'r--', linewidth=2)
plt.ylim([0, 10])
plt.legend(loc='lower right')
plt.title('Sepal Length vs Pedal Width')
plt.xlabel('Pedal Width')
plt.ylabel('Sepal Length')
plt.show()
plt.plot(train_loss, 'k-', label='Train Set Loss')
plt.plot(test_loss, 'r--', label='Test Set Loss')
plt.title('L2 Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('L2 Loss')
plt.legend(loc='upper right')
plt.show()[/mw_shl_code]

怎么运行
直觉上,我们可以将SVM回归看作是一个试图适应多个点的函数 尽可能宽的边距。 该线的拟合对该参数有些敏感。 如果我们选择太小的epsilon,算法将无法适应边距中的许多点。 如果我们选择太大的epsilon,将会有许多行能够适应边距中的所有数据点。 我们更喜欢较小的epsilon,因为距离边缘更近的点比距离更远的点贡献更少的损失。
在TensorFlow中使用内核
先前的SVM使用线性可分数据。 如果我们想要分离非线性数据,我们可以改变将线性分隔符投影到数据上的方式。 这是通过更改SVM丢失函数中的内核来完成的。 在本章中,我们将介绍如何更换内核并分离非线性可分离数据。
做好准备
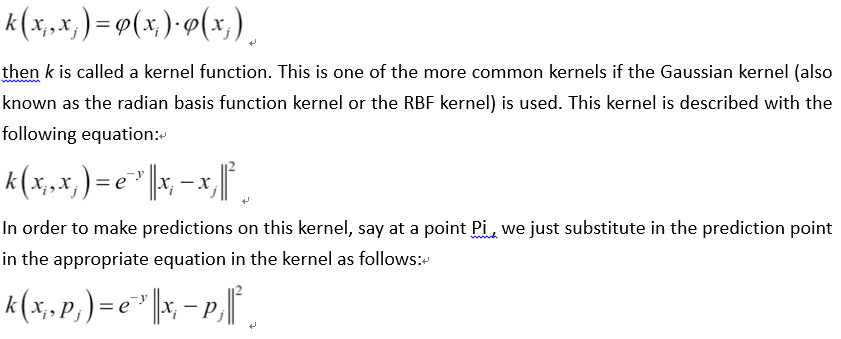
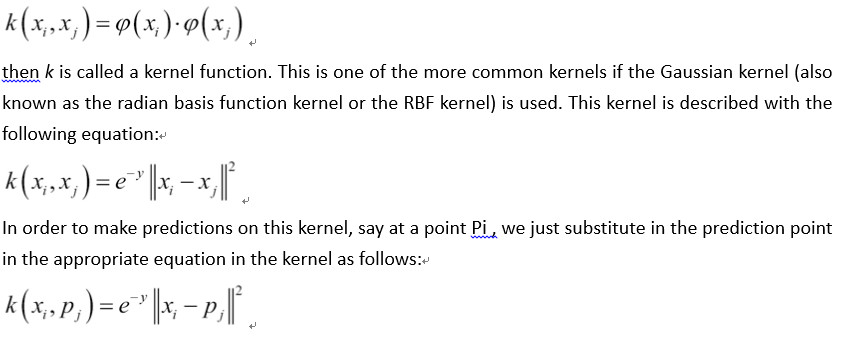
在本文中,我们将激励支持向量机中内核的使用。 在线性SVM部分,我们用特定的损失函数求解了软边界。 这种方法的另一种方法是解决所谓的优化问题的对偶。 可以证明线性SVM问题的对偶性由以下公式给出:
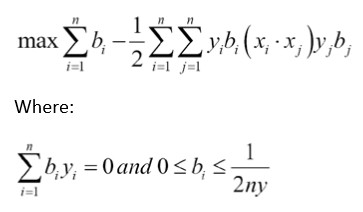
这里,模型中的变量将是b向量。 理想情况下,此向量将非常稀疏,仅对我们数据集的相应支持向量采用接近1和-1的值。 我们的数据点向量由Xi表示,我们的目标(1或-1)由Yi表示
前面等式中的内核是点积Xi * Xj,它给出了线性内核。 该内核是一个方形矩阵,填充了数据点的i,j点积。

在本节中,我们将讨论如何实现高斯内核。 我们还将记录在适当的地方替换实现线性内核的位置。 我们将使用的数据集将手动创建,以显示高斯内核更适合在线性内核上使用的位置。
怎么做
1.首先,我们加载必要的库并启动图形会话,如下所示:
[mw_shl_code=python,true]import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn import datasets
sess = tf.Session() [/mw_shl_code]
2.现在我们生成数据。 我们将生成的数据将是两个同心数据环,每个环将属于不同的类。 我们必须确保类只有-1或1。 然后我们将数据分成每个类的x和y值以用于绘图目的。 使用以下代码:
[mw_shl_code=python,true]
(x_vals, y_vals) = datasets.make_circles(n_samples=500, factor=.5, noise=.1)
y_vals = np.array([1 if y==1 else -1 for y in y_vals])
class1_x = [x[0] for i,x in enumerate(x_vals) if y_vals==1]
class1_y = [x[1] for i,x in enumerate(x_vals) if y_vals==1]
class2_x = [x[0] for i,x in enumerate(x_vals) if y_vals==-1]
class2_y = [x[1] for i,x in enumerate(x_vals) if y_vals==-1] [/mw_shl_code]
3.接下来我们声明我们的批量大小,占位符,并创建我们的模型变量,b。 对于SVM,我们倾向于需要更大的批量大小,因为我们需要一个非常稳定的模型,该模型在每次训练生成时都不会波动很大。 另请注意,我们为预测点添加了额外的占位符。 为了可视化结果,我们将创建一个颜色网格,以查看哪些区域最后属于哪个类。 使用以下代码:
[mw_shl_code=python,true]batch_size = 250
x_data = tf.placeholder(shape=[None, 2], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
prediction_grid = tf.placeholder(shape=[None, 2], dtype=tf. float32)
b = tf.Variable(tf.random_normal(shape=[1,batch_size])) [/mw_shl_code]
4.我们现在将创建高斯内核。 该内核可以表示为矩阵运算,如下所示:
[mw_shl_code=python,true]gamma = tf.constant(-50.0)
dist = tf.reduce_sum(tf.square(x_data), 1)
dist = tf.reshape(dist, [-1,1])
sq_dists = tf.add(tf.sub(dist, tf.mul(2., tf.matmul(x_data, tf.transpose(x_data)))), tf.transpose(dist))
my_kernel = tf.exp(tf.mul(gamma, tf.abs(sq_dists)))[/mw_shl_code]
请注意在加法和减法操作的sq_dists行中使用广播。注意,线性内核可以表示为
[mw_shl_code=python,true]my_kernel = tf.matmul(x_data,tf.transpose(x_data))[/mw_shl_code]
5.现在我们按照本文中的说明声明了双重问题。 最后,我们将使用tf.neg()函数最小化损失函数的负面,而不是最大化。 使用以下代码:
[mw_shl_code=python,true]model_output = tf.matmul(b, my_kernel)
first_term = tf.reduce_sum(b)
b_vec_cross = tf.matmul(tf.transpose(b), b)
y_target_cross = tf.matmul(y_target, tf.transpose(y_target))
second_term = tf.reduce_sum(tf.mul(my_kernel, tf.mul(b_vec_cross, y_target_cross)))
loss = tf.neg(tf.sub(first_term, second_term)) [/mw_shl_code]
6.我们现在创建预测和准确度函数。 首先,我们必须创建一个类似于步骤4的预测内核,但是我们拥有带有预测数据的点的核心,而不是点的内核。 然后预测是模型输出的符号。 使用以下代码:
[mw_shl_code=python,true]rA = tf.reshape(tf.reduce_sum(tf.square(x_data), 1),[-1,1])
rB = tf.reshape(tf.reduce_sum(tf.square(prediction_grid), 1),[- 1,1])
pred_sq_dist = tf.add(tf.sub(rA, tf.mul(2., tf.matmul(x_data, tf.transpose(prediction_grid)))), tf.transpose(rB))
pred_kernel = tf.exp(tf.mul(gamma, tf.abs(pred_sq_dist)))
prediction_output = tf.matmul(tf.mul(tf.transpose(y_target),b), pred_kernel)
prediction = tf.sign(prediction_output-tf.reduce_mean(prediction_ output))
accuracy = tf.reduce_mean(tf.cast(tf.equal(tf.squeeze(prediction), tf.squeeze(y_target)), tf.float32))[/mw_shl_code]
为了实现线性预测内核,我们可以编写
[mw_shl_code=python,true]pred_kernel = tf.matmul(x_data,tf.transpose(prediction_grid))[/mw_shl_code]
7.现在我们可以创建一个优化器函数并初始化所有变量,如下所示:
[mw_shl_code=python,true]my_opt = tf.train.GradientDescentOptimizer(0.001)
train_step = my_opt.minimize(loss)
init = tf.initialize_all_variables()
sess.run(init)
[/mw_shl_code]
8.接下来我们开始训练循环。 我们将记录每代的损耗矢量和批次精度。 当我们运行准确度时,我们必须放入所有三个占位符,但我们输入x数据两次以获得对点的预测。 使用以下代码:
[mw_shl_code=python,true]loss_vec = []
batch_accuracy = []
for i in range(500):
rand_index = np.random.choice(len(x_vals), size=batch_size)
rand_x = x_vals[rand_index]
rand_y = np.transpose([y_vals[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
temp_loss = sess.run(loss, feed_dict={x_data: rand_x, y_ target: rand_y})
loss_vec.append(temp_loss)
acc_temp = sess.run(accuracy, feed_dict={x_data: rand_x,
y_target: rand_y,
prediction_ grid:rand_x})
batch_accuracy.append(acc_temp)
if (i+1)%100==0:
print('Step #' + str(i+1))
print('Loss = ' + str(temp_loss))
[/mw_shl_code]
9.这导致以下输出:
[mw_shl_code=python,true]Step #100
Loss = -28.0772
Step #200
Loss = -3.3628
Step #300
Loss = -58.862
Step #400
Loss = -75.1121
Step #500
Loss = -84.8905[/mw_shl_code]
10.为了查看整个空间的输出类,我们将在系统中创建一个预测点网格,并对所有预测点进行预测,如下所示:
[mw_shl_code=python,true]x_min, x_max = x_vals[:, 0].min() - 1, x_vals[:, 0].max() + 1
y_min, y_max = x_vals[:, 1].min() - 1, x_vals[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
grid_points = np.c_[xx.ravel(), yy.ravel()]
[grid_predictions] = sess.run(prediction, feed_dict={x_data: rand_x,
y_target: rand_y,
prediction_ grid: grid_points})
grid_predictions = grid_predictions.reshape(xx.shape) [/mw_shl_code]
11.以下是绘制结果,批次准确度和损失的代码:
[mw_shl_code=python,true]plt.contourf(xx, yy, grid_predictions, cmap=plt.cm.Paired, alpha=0.8)
plt.plot(class1_x, class1_y, 'ro', label='Class 1')
plt.plot(class2_x, class2_y, 'kx', label='Class -1')
plt.legend(loc='lower right')
plt.ylim([-1.5, 1.5])
plt.xlim([-1.5, 1.5])
plt.show()
plt.plot(batch_accuracy, 'k-', label='Accuracy')
plt.title('Batch Accuracy')
plt.xlabel('Generation')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.show()
plt.plot(loss_vec, 'k-')
plt.title('Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('Loss')
plt.show()[/mw_shl_code]
12.为简洁起见,我们只显示结果图,但我们也可以单独运行绘图代码,如果我们选择的话,可以看到所有三个:
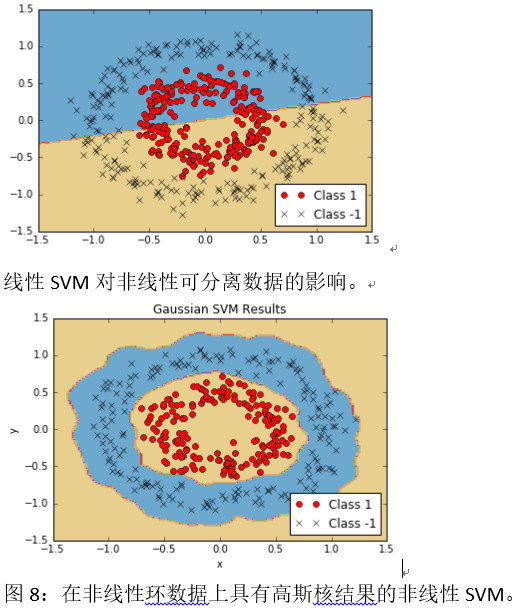
具有高斯核的非线性SVM导致非线性环数据。
怎么运行的
有两个重要的代码要知道:我们如何实现内核以及我们如何为SVM双优化问题实现损失函数。 我们已经展示了如何实现线性和高斯核,并且高斯核可以分离非线性数据集。
我们还应该提到另一个参数,即高斯核中的伽马值。 此参数控制影响点对分离曲率的影响程度。 通常选择小值,但它在很大程度上取决于数据集。 理想情况下,此参数是使用统计技术(如交叉验证)选择的。
还有更多
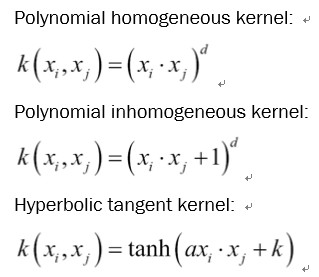
如果我们这样选择,我们可以实现更多内核。 以下是一些更常见的非线性内核列表:

原文:
Reduction to Linear Regression
Support vector machines can be used to fit linear regression. In this chapter, we will explore how to do this with TensorFlow.
Getting ready
The same maximum margin concept can be applied toward fitting linear regression. Instead of maximizing the margin that separates the classes, we can think about maximizing the margin that contains the most (x, y) points. To illustrate this, we will use the same iris data set, and show that we can use this concept to fit a line between sepal length and petal width.
The corresponding loss function will be similar to max
 Here, is half of the width of the margin, which makes the loss equal to zero if a point lies in this region.
Here, is half of the width of the margin, which makes the loss equal to zero if a point lies in this region.
How to do it…
1.First we load the necessary libraries, start a graph, and load the iris dataset. After that, we will split the dataset into train and test sets to visualize the loss on both. Use the following code:
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn import datasets
sess = tf.Session()
iris = datasets.load_iris()
x_vals = np.array([x[3] for x in iris.data])
y_vals = np.array([y[0] for y in iris.data])
train_indices = np.random.choice(len(x_vals), round(len(x_ vals)*0.8), replace=False)
test_indices = np.array(list(set(range(len(x_vals))) - set(train_ indices)))
x_vals_train = x_vals[train_indices]
x_vals_test = x_vals[test_indices]
y_vals_train = y_vals[train_indices]
y_vals_test = y_vals[test_indices]
For this example, we have split the data into train and test. It is also common to split the data into three datasets, which includes the validation set. We can use this validation set to verify that we are not overfitting models as we train them.
2.Let's declare our batch size, placeholders, and variables, and create our linear model, as follows:
batch_size = 50
x_data = tf.placeholder(shape=[None, 1], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
A = tf.Variable(tf.random_normal(shape=[1,1]))
b = tf.Variable(tf.random_normal(shape=[1,1]))
model_output = tf.add(tf.matmul(x_data, A), b)
3.Now we declare our loss function. The loss function, as described in the preceding text, is implemented to follow with . Remember that the epsilon is part of our loss function, which allows for a soft margin instead of a hard margin.
epsilon = tf.constant([0.5])
loss = tf.reduce_mean(tf.maximum(0., tf.sub(tf.abs(tf.sub(model_ output, y_target)), epsilon)))
4.We create an optimizer and initialize our variables next, as follows:
my_opt = tf.train.GradientDescentOptimizer(0.075)
train_step = my_opt.minimize(loss)
init = tf.initialize_all_variables()
sess.run(init)
5.Now we iterate through 200 training iterations and save the training and test loss for plotting later:
train_loss = []
test_loss = []
for i in range(200):
rand_index = np.random.choice(len(x_vals_train), size=batch_ size)
rand_x = np.transpose([x_vals_train[rand_index]])
rand_y = np.transpose([y_vals_train[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
temp_train_loss = sess.run(loss, feed_dict={x_data: np.transpose([x_vals_train]), y_target: np.transpose([y_vals_ train])})
train_loss.append(temp_train_loss)
temp_test_loss = sess.run(loss, feed_dict={x_data: np.transpose([x_vals_test]), y_target: np.transpose([y_vals_ test])})
test_loss.append(temp_test_loss)
if (i+1)%50==0:
print('-----------')
print('Generation: ' + str(i))
print('A = ' + str(sess.run(A)) + ' b = ' + str(sess. run(b)))
print('Train Loss = ' + str(temp_train_loss))
print('Test Loss = ' + str(temp_test_loss))
6.This results in the following output:
Generation: 50
A = [[ 2.20651722]] b = [[ 2.71290684]]
Train Loss = 0.609453
Test Loss = 0.460152
-----------
Generation: 100
A = [[ 1.6440177]] b = [[ 3.75240564]]
Train Loss = 0.242519
Test Loss = 0.208901
-----------
Generation: 150
A = [[ 1.27711761]] b = [[ 4.3149066]]
Train Loss = 0.108192
Test Loss = 0.119284
-----------
Generation: 200
A = [[ 1.05271816]] b = [[ 4.53690529]]
Train Loss = 0.0799957
Test Loss = 0.107551
7.We can now extract the coefficients we found, and get values for the best-fit line. For plotting purposes, we will also get values for the margins as well. Use the following code:
[[slope]] = sess.run(A)
[[y_intercept]] = sess.run(b)
[width] = sess.run(epsilon)
best_fit = []
best_fit_upper = []
best_fit_lower = []
for i in x_vals:
best_fit.append(slope*i+y_intercept)
best_fit_upper.append(slope*i+y_intercept+width)
best_fit_lower.append(slope*i+y_intercept-width)
8.Finally, here is the code to plot the data with the fitted line and the train-test loss:
plt.plot(x_vals, y_vals, 'o', label='Data Points')
plt.plot(x_vals, best_fit, 'r-', label='SVM Regression Line', linewidth=3)
plt.plot(x_vals, best_fit_upper, 'r--', linewidth=2)
plt.plot(x_vals, best_fit_lower, 'r--', linewidth=2)
plt.ylim([0, 10])
plt.legend(loc='lower right')
plt.title('Sepal Length vs Pedal Width')
plt.xlabel('Pedal Width')
plt.ylabel('Sepal Length')
plt.show()
plt.plot(train_loss, 'k-', label='Train Set Loss')
plt.plot(test_loss, 'r--', label='Test Set Loss')
plt.title('L2 Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('L2 Loss')
plt.legend(loc='upper right')
plt.show()
How it works…
Intuitively, we can think of SVM regression as a function that is trying to fit as many points in the width margin from the line as possible. The fitting of this line is somewhat sensitive to this parameter. If we choose too small an epsilon, the algorithm will not be able to fit many points in the margin. If we choose too large of an epsilon, there will be many lines that are able to fit all the data points in the margin. We prefer a smaller epsilon, since nearer points to the margin contribute less loss than further away points.
Working with Kernels in TensorFlow
The prior SVMs worked with linear separable data. If we would like to separate non-linear data, we can change how we project the linear separator onto the data. This is done by changing the kernel in the SVM loss function. In this chapter, we introduce how to changer kernels and separate non-linear separable data.
Getting ready
In this recipe, we will motivate the usage of kernels in support vector machines. In the linear SVM section, we solved the soft margin with a specific loss function. A different approach to this method is to solve what is called the dual of the optimization problem. It can be shown that the dual for the linear SVM problem is given by the following formula:
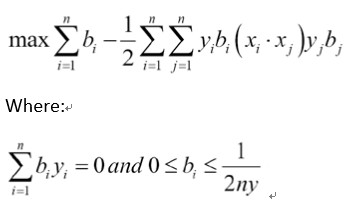
Here, the variable in the model will be the b vector. Ideally, this vector will be quite sparse, only taking on values near 1 and -1 for the corresponding support vectors of our dataset. Our data point vectors are indicated by Xi and our targets (1 or -1) are represented by Yi
The kernel in the preceding equations is the dot product Xi*Xj , which gives us the linear kernel. This kernel is a square matrix filled with the i,j dot products of the data points.
In this section, we will discuss how to implement the Gaussian kernel. We will also make a note of where to make the substitution for implementing the linear kernel where appropriate. The dataset we will use will be manually created to show where the Gaussian kernel would be more appropriate to use over the linear kernel.
How to do it…
1.First we load the necessary libraries and start a graph session, as follows:
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn import datasets
sess = tf.Session()
2.Now we generate the data. The data we will generate will be two concentric rings of data, each ring will belong to a different class. We have to make sure that the classes are -1 or 1 only . Then we will split the data into x and y values for each class for plotting purposes. Use the following code:
(x_vals, y_vals) = datasets.make_circles(n_samples=500, factor=.5, noise=.1)
y_vals = np.array([1 if y==1 else -1 for y in y_vals])
class1_x = [x[0] for i,x in enumerate(x_vals) if y_vals==1]
class1_y = [x[1] for i,x in enumerate(x_vals) if y_vals==1]
class2_x = [x[0] for i,x in enumerate(x_vals) if y_vals==-1]
class2_y = [x[1] for i,x in enumerate(x_vals) if y_vals==-1]
3.Next we declare our batch size, placeholders, and create our model variable, b. For SVMs we tend to want larger batch sizes because we want a very stable model that won't fluctuate much with each training generation. Also note that we have an extra placeholder for the prediction points. To visualize the results, we will create a color grid to see which areas belong to which class at the end. Use the following code:
batch_size = 250
x_data = tf.placeholder(shape=[None, 2], dtype=tf.float32)
y_target = tf.placeholder(shape=[None, 1], dtype=tf.float32)
prediction_grid = tf.placeholder(shape=[None, 2], dtype=tf. float32)
b = tf.Variable(tf.random_normal(shape=[1,batch_size]))
4.We will now create the Gaussian kernel. This kernel can be expressed as matrix operations as follows:
gamma = tf.constant(-50.0)
dist = tf.reduce_sum(tf.square(x_data), 1)
dist = tf.reshape(dist, [-1,1])
sq_dists = tf.add(tf.sub(dist, tf.mul(2., tf.matmul(x_data, tf.transpose(x_data)))), tf.transpose(dist))
my_kernel = tf.exp(tf.mul(gamma, tf.abs(sq_dists)))
Note the usage of broadcasting in the sq_dists line of the add and subtract operations.
Note that the linear kernel can be expressed as my_kernel = tf.matmul(x_data, tf.transpose(x_data)).
5.Now we declare the dual problem as previously stated in this recipe. At the end, instead of maximizing, we will be minimizing the negative of the loss function with a tf.neg() function. Use the following code:
model_output = tf.matmul(b, my_kernel)
first_term = tf.reduce_sum(b)
b_vec_cross = tf.matmul(tf.transpose(b), b)
y_target_cross = tf.matmul(y_target, tf.transpose(y_target))
second_term = tf.reduce_sum(tf.mul(my_kernel, tf.mul(b_vec_cross, y_target_cross)))
loss = tf.neg(tf.sub(first_term, second_term))
6.We now create the prediction and accuracy functions. First, we must create a prediction kernel, similar to step 4, but instead of a kernel of the points with itself, we have the kernel of the points with the prediction data. The prediction is then the sign of the output of the model. Use the following code:
rA = tf.reshape(tf.reduce_sum(tf.square(x_data), 1),[-1,1])
rB = tf.reshape(tf.reduce_sum(tf.square(prediction_grid), 1),[- 1,1])
pred_sq_dist = tf.add(tf.sub(rA, tf.mul(2., tf.matmul(x_data, tf.transpose(prediction_grid)))), tf.transpose(rB))
pred_kernel = tf.exp(tf.mul(gamma, tf.abs(pred_sq_dist)))
prediction_output = tf.matmul(tf.mul(tf.transpose(y_target),b), pred_kernel)
prediction = tf.sign(prediction_output-tf.reduce_mean(prediction_ output))
accuracy = tf.reduce_mean(tf.cast(tf.equal(tf.squeeze(prediction), tf.squeeze(y_target)), tf.float32))
To implement the linear prediction kernel, we can write pred_kernel = tf.matmul(x_data, tf.transpose(prediction_grid)).
7.Now we can create an optimizer function and initialize all the variables, as follows:
my_opt = tf.train.GradientDescentOptimizer(0.001)
train_step = my_opt.minimize(loss)
init = tf.initialize_all_variables()
sess.run(init)
8.Next we start the training loop. We will record the loss vector and the batch accuracy for each generation. When we run the accuracy, we have to put in all three placeholders, but we feed in the x data twice to get the prediction on the points. Use the following code:
loss_vec = []
batch_accuracy = []
for i in range(500):
rand_index = np.random.choice(len(x_vals), size=batch_size)
rand_x = x_vals[rand_index]
rand_y = np.transpose([y_vals[rand_index]])
sess.run(train_step, feed_dict={x_data: rand_x, y_target: rand_y})
temp_loss = sess.run(loss, feed_dict={x_data: rand_x, y_ target: rand_y})
loss_vec.append(temp_loss)
acc_temp = sess.run(accuracy, feed_dict={x_data: rand_x,
y_target: rand_y,
prediction_ grid:rand_x})
batch_accuracy.append(acc_temp)
if (i+1)%100==0:
print('Step #' + str(i+1))
print('Loss = ' + str(temp_loss))
9.This results in the following output:
Step #100
Loss = -28.0772
Step #200
Loss = -3.3628
Step #300
Loss = -58.862
Step #400
Loss = -75.1121
Step #500
Loss = -84.8905
10.In order to see the output class on the whole space, we will create a mesh of prediction points in our system and run the prediction on all of them, as follows:
x_min, x_max = x_vals[:, 0].min() - 1, x_vals[:, 0].max() + 1
y_min, y_max = x_vals[:, 1].min() - 1, x_vals[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
grid_points = np.c_[xx.ravel(), yy.ravel()]
[grid_predictions] = sess.run(prediction, feed_dict={x_data: rand_x,
y_target: rand_y,
prediction_ grid: grid_points})
grid_predictions = grid_predictions.reshape(xx.shape)
11.The following is the code to plot the result, batch accuracy, and loss:
plt.contourf(xx, yy, grid_predictions, cmap=plt.cm.Paired, alpha=0.8)
plt.plot(class1_x, class1_y, 'ro', label='Class 1')
plt.plot(class2_x, class2_y, 'kx', label='Class -1')
plt.legend(loc='lower right')
plt.ylim([-1.5, 1.5])
plt.xlim([-1.5, 1.5])
plt.show()
plt.plot(batch_accuracy, 'k-', label='Accuracy')
plt.title('Batch Accuracy')
plt.xlabel('Generation')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.show()
plt.plot(loss_vec, 'k-')
plt.title('Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('Loss')
plt.show()
12.For succinctness, we will show only the results graph, but we can also separately run the plotting code and see all three if we so choose:
Non-linear SVM with Gaussian kernel results on nonlinear ring data.
How it works…
There are two important pieces of the code to know about: how we implemented the kernel and how we implemented the loss function for the SVM dual optimization problem. We have shown how to implement the linear and Gaussian kernel and that the Gaussian kernel can separate nonlinear datasets.
We should also mention that there is another parameter, the gamma value in the Gaussian kernel. This parameter controls how much influence points have on the curvature of the separation. Small values are commonly chosen, but it depends heavily on the dataset. Ideally this parameter is chosen with statistical techniques such as cross-validation.
There's more…
There are many more kernels that we could implement if we so choose. Here is a list of a few more common nonlinear kernels:
|
 /2
/2 